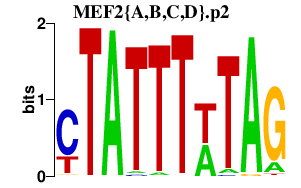
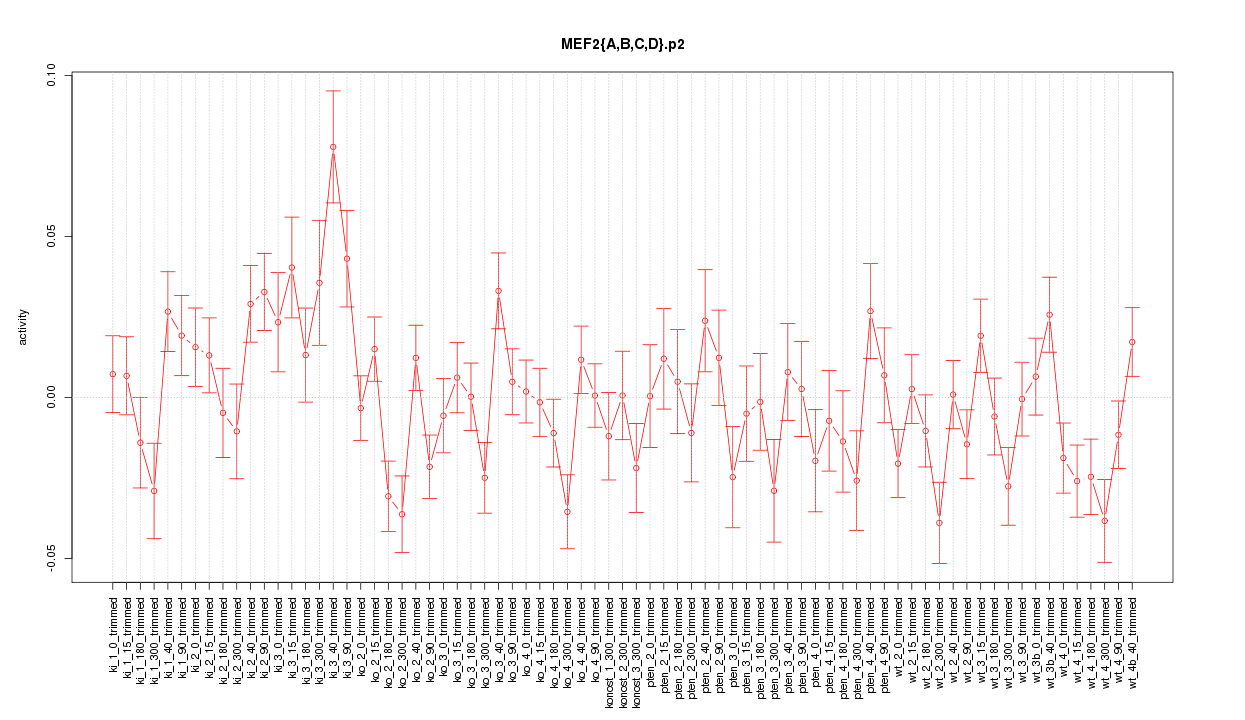
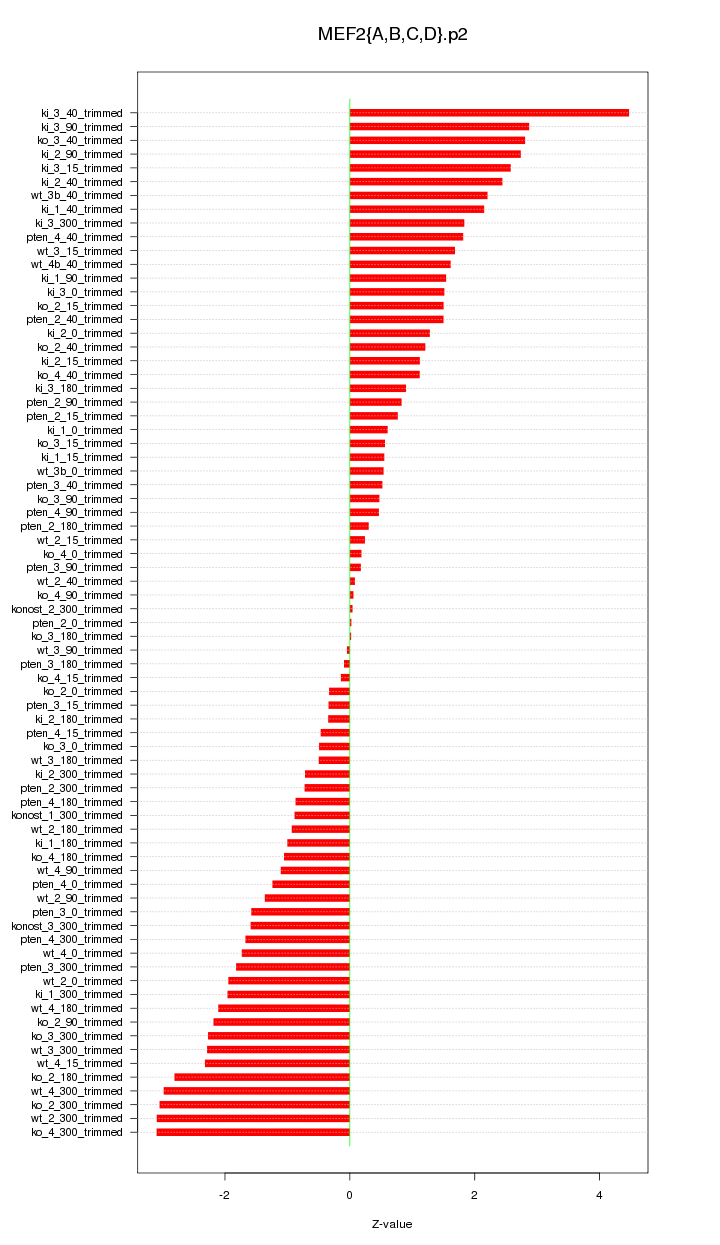
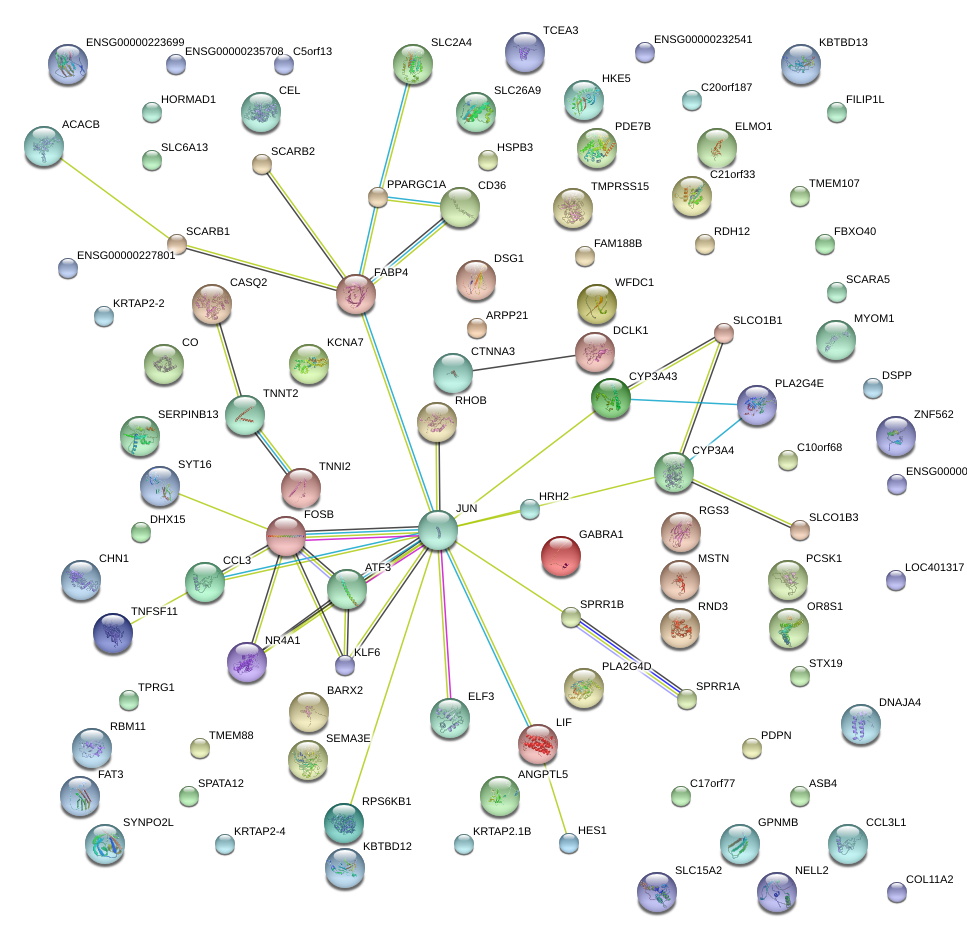
|
chr12_+_52445185
|
43.696
|
NM_002135
NM_173157
|
NR4A1
|
nuclear receptor subfamily 4, group A, member 1
|
|
chr7_+_28448970
|
12.280
|
|
LOC401317
|
uncharacterized LOC401317
|
|
chr12_-_45269340
|
12.188
|
NM_001145109
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr11_+_129245834
|
9.626
|
NM_003658
|
BARX2
|
BARX homeobox 2
|
|
chr19_+_45971249
|
9.441
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr1_+_201979747
|
8.844
|
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr17_+_7758380
|
7.680
|
NM_203411
|
TMEM88
|
transmembrane protein 88
|
|
chr4_-_23891608
|
7.621
|
NM_013261
|
PPARGC1A
|
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha
|
|
chr8_-_82395401
|
7.148
|
NM_001442
|
FABP4
|
fatty acid binding protein 4, adipocyte
|
|
chr1_+_201979689
|
6.853
|
NM_001114309
NM_004433
|
ELF3
|
E74-like factor 3 (ets domain transcription factor, epithelial-specific )
|
|
chr6_+_136172802
|
6.720
|
NM_018945
|
PDE7B
|
phosphodiesterase 7B
|
|
chr5_+_53751430
|
6.381
|
NM_006308
|
HSPB3
|
heat shock 27kDa protein 3
|
|
chr17_-_39211462
|
5.699
|
NM_033032
|
KRTAP2-2
|
keratin associated protein 2-2
|
|
chr2_+_20646831
|
5.577
|
NM_004040
|
RHOB
|
ras homolog gene family, member B
|
|
chr5_-_111091947
|
5.526
|
NM_001142483
|
C5orf13
|
chromosome 5 open reading frame 13
|
|
chr7_+_23286390
|
5.500
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr13_+_43148182
|
5.313
|
NM_003701
|
TNFSF11
|
tumor necrosis factor (ligand) superfamily, member 11
|
|
chr10_+_32856650
|
5.225
|
NM_024688
|
C10orf68
|
chromosome 10 open reading frame 68
|
|
chr1_-_59249687
|
5.172
|
NM_002228
|
JUN
|
jun proto-oncogene
|
|
chr3_-_93747453
|
4.792
|
NM_001001850
|
STX19
|
syntaxin 19
|
|
chr18_+_28898051
|
4.744
|
NM_001942
|
DSG1
|
desmoglein 1
|
|
chr19_+_20959109
|
4.687
|
|
|
|
|
chr15_+_78558522
|
4.673
|
NM_001130183
|
DNAJA4
|
DnaJ (Hsp40) homolog, subfamily A, member 4
|
|
chr3_+_57094468
|
4.623
|
NM_181727
|
SPATA12
|
spermatogenesis associated 12
|
|
chr1_+_152956563
|
4.575
|
NM_001199828
NM_005987
|
SPRR1A
|
small proline-rich protein 1A
|
|
chr11_+_1860232
|
4.502
|
NM_003282
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr5_-_95748688
|
4.421
|
NM_001177876
|
PCSK1
|
proprotein convertase subtilisin/kexin type 1
|
|
chr7_+_23286386
|
4.336
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr17_+_7184978
|
4.112
|
NM_001042
|
SLC2A4
|
solute carrier family 2 (facilitated glucose transporter), member 4
|
|
chr11_+_1860718
|
4.094
|
NM_001145829
|
TNNI2
|
troponin I type 2 (skeletal, fast)
|
|
chr10_-_69425356
|
4.038
|
NM_001127384
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr10_-_75415748
|
4.028
|
NM_001114133
|
SYNPO2L
|
synaptopodin 2-like
|
|
chr12_-_45270157
|
3.975
|
NM_001145108
NM_006159
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr1_+_212782103
|
3.933
|
|
ATF3
|
activating transcription factor 3
|
|
chr14_+_62462540
|
3.928
|
NM_031914
|
SYT16
|
synaptotagmin XVI
|
|
chr3_+_35683848
|
3.828
|
NM_016300
|
ARPP21
|
cAMP-regulated phosphoprotein, 21kDa
|
|
chr1_+_13911966
|
3.711
|
NM_001006624
NM_001006625
|
PDPN
|
podoplanin
|
|
chr4_+_41362843
|
3.686
|
|
|
|
|
chr15_+_65369153
|
3.669
|
NM_001101362
|
KBTBD13
|
kelch repeat and BTB (POZ) domain containing 13
|
|
chr3_-_99833348
|
3.650
|
NM_001042459
NM_182909
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr1_-_205912546
|
3.648
|
NM_052934
NM_134325
|
SLC26A9
|
solute carrier family 26, member 9
|
|
chr2_-_190927321
|
3.582
|
NM_005259
|
MSTN
|
myostatin
|
|
chr17_-_34524054
|
3.578
|
NM_001001437
NM_021006
|
CCL3L3
CCL3
CCL3L1
|
chemokine (C-C motif) ligand 3-like 3
chemokine (C-C motif) ligand 3
chemokine (C-C motif) ligand 3-like 1
|
|
chr12_+_48919414
|
3.531
|
NM_001005203
|
OR8S1
|
olfactory receptor, family 8, subfamily S, member 1
|
|
chr7_+_23286420
|
3.515
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr1_-_201346804
|
3.505
|
NM_000364
NM_001001430
NM_001001431
NM_001001432
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr12_+_20963637
|
3.498
|
NM_019844
|
SLCO1B3
|
solute carrier organic anion transporter family, member 1B3
|
|
chr1_-_116311398
|
3.476
|
NM_001232
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr12_-_45270071
|
3.473
|
|
NELL2
|
NEL-like 2 (chicken)
|
|
chr9_+_116356407
|
3.469
|
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr15_-_42386657
|
3.467
|
NM_178034
|
PLA2G4D
|
phospholipase A2, group IVD (cytosolic)
|
|
chr8_-_27850368
|
3.466
|
NM_173833
|
SCARA5
|
scavenger receptor class A, member 5 (putative)
|
|
chr3_+_188664939
|
3.460
|
|
TPRG1
|
tumor protein p63 regulated 1
|
|
chr7_+_23286401
|
3.388
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr18_-_3219846
|
3.387
|
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr3_+_193853930
|
3.332
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_+_121312169
|
3.285
|
NM_016298
|
FBXO40
|
F-box protein 40
|
|
chr7_-_99381703
|
3.282
|
NM_001202855
NM_017460
|
CYP3A4
|
cytochrome P450, family 3, subfamily A, polypeptide 4
|
|
chr3_+_127634317
|
3.252
|
|
KBTBD12
|
kelch repeat and BTB (POZ) domain containing 12
|
|
chr1_-_23751126
|
3.203
|
NM_003196
|
TCEA3
|
transcription elongation factor A (SII), 3
|
|
chr1_-_116311330
|
3.196
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr12_-_371994
|
3.192
|
NM_001190997
NM_001243392
NM_016615
|
SLC6A13
|
solute carrier family 6 (neurotransmitter transporter, GABA), member 13
|
|
chr18_+_61254533
|
3.180
|
NM_012397
|
SERPINB13
|
serpin peptidase inhibitor, clade B (ovalbumin), member 13
|
|
chr14_+_68168602
|
3.157
|
NM_152443
|
RDH12
|
retinol dehydrogenase 12 (all-trans/9-cis/11-cis)
|
|
chr21_+_15588465
|
3.154
|
NM_144770
|
RBM11
|
RNA binding motif protein 11
|
|
chr22_-_30642683
|
3.151
|
NM_002309
|
LIF
|
leukemia inhibitory factor (cholinergic differentiation factor)
|
|
chr1_-_116311161
|
3.134
|
|
CASQ2
|
calsequestrin 2 (cardiac muscle)
|
|
chr12_+_21284127
|
3.129
|
NM_006446
|
SLCO1B1
|
solute carrier organic anion transporter family, member 1B1
|
|
chr11_+_92085261
|
3.105
|
NM_001008781
|
FAT3
|
FAT tumor suppressor homolog 3 (Drosophila)
|
|
chr12_+_109554391
|
3.102
|
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr16_+_84328400
|
3.071
|
NM_021197
|
WFDC1
|
WAP four-disulfide core domain 1
|
|
chr7_+_95115212
|
3.064
|
NM_016116
NM_145872
|
ASB4
|
ankyrin repeat and SOCS box containing 4
|
|
chr9_-_102582143
|
3.021
|
|
|
|
|
chr2_-_175712276
|
3.016
|
NM_001206602
|
CHN1
|
chimerin (chimaerin) 1
|
|
chr1_-_150693321
|
2.975
|
NM_001199829
NM_032132
|
HORMAD1
|
HORMA domain containing 1
|
|
chr15_-_42342900
|
2.971
|
NM_001206670
|
PLA2G4E
|
phospholipase A2, group IVE
|
|
chr9_+_135937364
|
2.971
|
NM_001807
|
CEL
|
carboxyl ester lipase (bile salt-stimulated lipase)
|
|
chr11_-_101787252
|
2.957
|
NM_178127
|
ANGPTL5
|
angiopoietin-like 5
|
|
chr2_-_151344145
|
2.876
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr7_+_99425635
|
2.871
|
NM_022820
NM_057095
NM_057096
|
CYP3A43
|
cytochrome P450, family 3, subfamily A, polypeptide 43
|
|
chr5_+_161275185
|
2.871
|
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr6_-_33160199
|
2.868
|
NM_001163771
NM_080679
NM_080680
NM_080681
|
COL11A2
|
collagen, type XI, alpha 2
|
|
chr4_+_88529680
|
2.863
|
NM_014208
|
DSPP
|
dentin sialophosphoprotein
|
|
chr7_-_83278478
|
2.861
|
NM_012431
|
SEMA3E
|
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E
|
|
chr10_-_3827418
|
2.845
|
NM_001160124
NM_001160125
NM_001300
|
KLF6
|
Kruppel-like factor 6
|
|
chr5_+_175108463
|
2.833
|
NM_022304
|
HRH2
|
histamine receptor H2
|
|
chr3_+_121613170
|
2.825
|
NM_001145998
NM_021082
|
SLC15A2
|
solute carrier family 15 (H+/peptide transporter), member 2
|
|
chr7_+_80231503
|
2.800
|
NM_001001547
|
CD36
|
CD36 molecule (thrombospondin receptor)
|
|
chr19_-_49575878
|
2.796
|
|
KCNA7
|
potassium voltage-gated channel, shaker-related subfamily, member 7
|
|
chr21_-_19775958
|
2.790
|
NM_002772
|
TMPRSS15
|
transmembrane protease, serine 15
|
|
chr6_+_27292539
|
2.782
|
NM_032030
|
FKSG83
|
FKSG83
|
|
chr17_+_72581056
|
2.780
|
NM_152460
|
C17orf77
|
chromosome 17 open reading frame 77
|
|
chr7_+_30951414
|
2.779
|
NM_198098
|
AQP1
|
aquaporin 1 (Colton blood group)
|
|
chr7_-_37488554
|
2.777
|
NM_014800
|
ELMO1
|
engulfment and cell motility 1
|
|
chr13_-_36429798
|
2.776
|
NM_001195415
NM_001195416
NM_001195430
|
DCLK1
|
doublecortin-like kinase 1
|
|
chr10_-_3827205
|
2.758
|
|
KLF6
|
Kruppel-like factor 6
|
|
chrX_-_18690187
|
2.756
|
NM_000330
|
RS1
|
retinoschisin 1
|
|
chr7_+_23286384
|
2.755
|
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr12_+_15699544
|
2.746
|
|
PTPRO
|
protein tyrosine phosphatase, receptor type, O
|
|
chr2_+_169921298
|
2.738
|
NM_005771
|
DHRS9
|
dehydrogenase/reductase (SDR family) member 9
|
|
chr10_+_91152321
|
2.732
|
NM_001548
|
IFIT1
|
interferon-induced protein with tetratricopeptide repeats 1
|
|
chr10_-_14879967
|
2.730
|
NM_001029954
|
CDNF
|
cerebral dopamine neurotrophic factor
|
|
chr19_-_50666279
|
2.726
|
NM_152358
|
IZUMO2
|
IZUMO family member 2
|
|
chr7_-_15726273
|
2.726
|
NM_005924
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr1_+_160051343
|
2.715
|
NM_004983
|
KCNJ9
|
potassium inwardly-rectifying channel, subfamily J, member 9
|
|
chr20_+_44657858
|
2.712
|
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr10_+_96443250
|
2.710
|
NM_000772
NM_001128925
|
CYP2C18
|
cytochrome P450, family 2, subfamily C, polypeptide 18
|
|
chr12_-_70004941
|
2.690
|
NM_201550
|
LRRC10
|
leucine rich repeat containing 10
|
|
chr1_+_70032810
|
2.686
|
|
|
|
|
chr7_+_23286277
|
2.682
|
NM_001005340
NM_002510
|
GPNMB
|
glycoprotein (transmembrane) nmb
|
|
chr7_-_30739660
|
2.677
|
NM_001202475
NM_001202481
|
CRHR2
|
corticotropin releasing hormone receptor 2
|
|
chr20_+_44657812
|
2.670
|
NM_020708
|
SLC12A5
|
solute carrier family 12 (potassium/chloride transporter), member 5
|
|
chr11_-_105010460
|
2.662
|
NM_021571
|
CARD18
|
caspase recruitment domain family, member 18
|
|
chr19_+_15904760
|
2.654
|
NM_001004466
|
OR10H5
|
olfactory receptor, family 10, subfamily H, member 5
|
|
chr6_+_26104103
|
2.634
|
NM_003542
|
HIST2H4B
HIST1H4C
|
histone cluster 2, H4b
histone cluster 1, H4c
|
|
chr2_+_233497964
|
2.591
|
|
EFHD1
|
EF-hand domain family, member D1
|
|
chrX_-_128657440
|
2.563
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr8_-_33457491
|
2.556
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr5_+_161274939
|
2.556
|
NM_001127644
|
GABRA1
|
gamma-aminobutyric acid (GABA) A receptor, alpha 1
|
|
chr3_-_42843217
|
2.555
|
|
|
|
|
chr1_-_152131703
|
2.548
|
NM_001122965
|
RPTN
|
repetin
|
|
chr7_+_18126543
|
2.547
|
NM_001204144
|
HDAC9
|
histone deacetylase 9
|
|
chr11_-_19223516
|
2.533
|
NM_001127656
NM_003476
|
CSRP3
|
cysteine and glycine-rich protein 3 (cardiac LIM protein)
|
|
chr12_-_22094166
|
2.526
|
|
ABCC9
|
ATP-binding cassette, sub-family C (CFTR/MRP), member 9
|
|
chr11_+_67171379
|
2.519
|
|
TBC1D10C
|
TBC1 domain family, member 10C
|
|
chr19_-_50832633
|
2.518
|
NM_004977
|
KCNC3
|
potassium voltage-gated channel, Shaw-related subfamily, member 3
|
|
chr3_+_42727010
|
2.501
|
NM_152393
|
KBTBD5
|
kelch repeat and BTB (POZ) domain containing 5
|
|
chr6_+_153019029
|
2.499
|
NM_025107
|
MYCT1
|
myc target 1
|
|
chrX_-_128657453
|
2.490
|
NM_003069
NM_139035
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr1_-_116383323
|
2.478
|
NM_001111061
|
NHLH2
|
nescient helix loop helix 2
|
|
chr6_+_152128453
|
2.472
|
NM_001122740
|
ESR1
|
estrogen receptor 1
|
|
chrX_-_128657381
|
2.459
|
|
SMARCA1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1
|
|
chr12_+_109665175
|
2.454
|
|
ACACB
|
acetyl-CoA carboxylase beta
|
|
chr19_-_20607761
|
2.447
|
|
ZNF826P
|
zinc finger protein 826, pseudogene
|
|
chr3_+_186714335
|
2.446
|
|
|
|
|
chr1_-_201346783
|
2.444
|
|
TNNT2
|
troponin T type 2 (cardiac)
|
|
chr7_-_15726026
|
2.426
|
|
MEOX2
|
mesenchyme homeobox 2
|
|
chr8_-_70745624
|
2.422
|
NM_001146008
|
SLCO5A1
|
solute carrier organic anion transporter family, member 5A1
|
|
chr1_+_160160284
|
2.418
|
NM_001231
|
CASQ1
|
calsequestrin 1 (fast-twitch, skeletal muscle)
|
|
chr6_+_152128685
|
2.416
|
NM_000125
|
ESR1
|
estrogen receptor 1
|
|
chr19_-_51487070
|
2.413
|
NM_139277
NM_001207053
NM_001243126
NM_005046
|
KLK7
|
kallikrein-related peptidase 7
|
|
chr3_-_39234071
|
2.411
|
NM_194293
NM_001198621
|
XIRP1
|
xin actin-binding repeat containing 1
|
|
chr9_+_132083294
|
2.398
|
NM_001012715
|
C9orf106
|
chromosome 9 open reading frame 106
|
|
chr8_-_33457279
|
2.376
|
|
DUSP26
|
dual specificity phosphatase 26 (putative)
|
|
chr9_+_75136716
|
2.373
|
NM_138691
|
TMC1
|
transmembrane channel-like 1
|
|
chr4_-_65275000
|
2.369
|
NM_001010874
|
TECRL
|
trans-2,3-enoyl-CoA reductase-like
|
|
chr2_+_204571197
|
2.364
|
NM_001243077
NM_001243078
NM_006139
|
CD28
|
CD28 molecule
|
|
chr19_-_54311958
|
2.353
|
NM_033297
|
NLRP12
|
NLR family, pyrin domain containing 12
|
|
chr21_+_19289656
|
2.348
|
NM_001204175
NM_001204176
NM_001204177
NM_001204178
|
CHODL
|
chondrolectin
|
|
chr5_+_34839268
|
2.344
|
NM_144725
|
TTC23L
|
tetratricopeptide repeat domain 23-like
|
|
chr3_-_46249787
|
2.336
|
NM_001295
|
CCR1
|
chemokine (C-C motif) receptor 1
|
|
chr2_+_85921413
|
2.326
|
NM_006433
NM_012483
|
GNLY
|
granulysin
|
|
chr3_+_193853937
|
2.325
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr3_-_152058778
|
2.313
|
NM_001123228
|
TMEM14E
|
transmembrane protein 14E
|
|
chr1_+_160085519
|
2.310
|
NM_000702
|
ATP1A2
|
ATPase, Na+/K+ transporting, alpha 2 polypeptide
|
|
chr1_-_115238091
|
2.308
|
NM_000036
NM_001172626
|
AMPD1
|
adenosine monophosphate deaminase 1
|
|
chr7_-_37488408
|
2.306
|
|
ELMO1
|
engulfment and cell motility 1
|
|
chr13_-_45775174
|
2.300
|
NM_198404
|
KCTD4
|
potassium channel tetramerisation domain containing 4
|
|
chr8_+_49984902
|
2.297
|
NM_001007176
|
C8orf22
|
chromosome 8 open reading frame 22
|
|
chr10_+_71561687
|
2.294
|
|
COL13A1
|
collagen, type XIII, alpha 1
|
|
chr19_+_23299776
|
2.291
|
|
ZNF730
|
zinc finger protein 730
|
|
chr8_+_55528626
|
2.289
|
NM_006269
|
RP1
|
retinitis pigmentosa 1 (autosomal dominant)
|
|
chr17_+_34087915
|
2.287
|
NM_145272
|
C17orf50
|
chromosome 17 open reading frame 50
|
|
chr1_-_27339326
|
2.274
|
NM_052943
|
FAM46B
|
family with sequence similarity 46, member B
|
|
chr22_-_38577692
|
2.273
|
NM_001004426
NM_003560
NM_001199562
|
PLA2G6
|
phospholipase A2, group VI (cytosolic, calcium-independent)
|
|
chr2_+_166428845
|
2.268
|
NM_024969
|
CSRNP3
|
cysteine-serine-rich nuclear protein 3
|
|
chr3_+_132036210
|
2.267
|
NM_001099
NM_001134194
|
ACPP
|
acid phosphatase, prostate
|
|
chr7_+_155250705
|
2.264
|
NM_001427
|
EN2
|
engrailed homeobox 2
|
|
chr3_+_108541544
|
2.264
|
NM_016388
|
TRAT1
|
T cell receptor associated transmembrane adaptor 1
|
|
chr18_-_3219986
|
2.261
|
NM_003803
NM_019856
|
MYOM1
|
myomesin 1, 185kDa
|
|
chr5_+_191625
|
2.259
|
NM_001080478
|
LRRC14B
|
leucine rich repeat containing 14B
|
|
chr1_-_152061529
|
2.258
|
NM_001008536
|
TCHHL1
|
trichohyalin-like 1
|
|
chr3_-_192445344
|
2.251
|
NM_004113
|
FGF12
|
fibroblast growth factor 12
|
|
chr16_+_12058963
|
2.243
|
NM_001192
|
TNFRSF17
|
tumor necrosis factor receptor superfamily, member 17
|
|
chr12_-_131200818
|
2.239
|
|
RIMBP2
|
RIMS binding protein 2
|
|
chr17_+_1665306
|
2.238
|
|
SERPINF1
|
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1
|
|
chr9_+_116356199
|
2.234
|
NM_144489
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_-_108231122
|
2.217
|
NM_001079874
|
VAV3
|
vav 3 guanine nucleotide exchange factor
|
|
chr18_-_35145999
|
2.215
|
NM_001025087
NM_001025088
NM_001025089
NM_020180
|
CELF4
|
CUGBP, Elav-like family member 4
|
|
chr10_-_69455926
|
2.213
|
NM_013266
|
CTNNA3
|
catenin (cadherin-associated protein), alpha 3
|
|
chr3_-_101039403
|
2.212
|
NM_016247
|
IMPG2
|
interphotoreceptor matrix proteoglycan 2
|
|
chr12_-_6756597
|
2.195
|
|
ACRBP
|
acrosin binding protein
|
|
chr19_+_10959105
|
2.190
|
NM_001136482
|
C19orf38
|
chromosome 19 open reading frame 38
|
|
chr9_+_139871954
|
2.188
|
NM_000954
|
PTGDS
|
prostaglandin D2 synthase 21kDa (brain)
|
|
chr2_-_217560247
|
2.185
|
NM_000599
|
IGFBP5
|
insulin-like growth factor binding protein 5
|
|
chr13_-_103411421
|
2.181
|
NM_001146197
|
CCDC168
|
coiled-coil domain containing 168
|
|
chr4_-_155312448
|
2.154
|
NM_017639
|
DCHS2
|
dachsous 2 (Drosophila)
|
|
chr8_+_67039130
|
2.154
|
NM_033058
NM_184085
NM_184086
NM_184087
|
TRIM55
|
tripartite motif containing 55
|
|
chr9_-_130540911
|
2.153
|
NM_170600
|
SH2D3C
|
SH2 domain containing 3C
|
|
chr3_-_108248168
|
2.152
|
NM_014981
|
MYH15
|
myosin, heavy chain 15
|
|
chr8_-_74171736
|
2.151
|
NM_001243237
|
LOC100130301
|
uncharacterized LOC100130301
|
|
chr17_+_7554711
|
2.140
|
|
ATP1B2
|
ATPase, Na+/K+ transporting, beta 2 polypeptide
|
|
chr19_+_52848681
|
2.134
|
NM_173530
|
ZNF610
|
zinc finger protein 610
|
|
chr1_+_152486977
|
2.134
|
NM_019060
|
CRCT1
|
cysteine-rich C-terminal 1
|
|
chr3_+_113616317
|
2.129
|
NM_001172105
|
GRAMD1C
|
GRAM domain containing 1C
|
|
chrX_-_63450486
|
2.120
|
NM_130388
|
ASB12
|
ankyrin repeat and SOCS box containing 12
|
|
chr9_+_103947316
|
2.117
|
NM_017753
|
LPPR1
|
lipid phosphate phosphatase-related protein type 1
|
|
chr5_+_140552201
|
2.116
|
NM_018940
|
PCDHB7
|
protocadherin beta 7
|
|
chr17_-_72855884
|
2.114
|
NM_000835
|
GRIN2C
|
glutamate receptor, ionotropic, N-methyl D-aspartate 2C
|
|
chr2_+_204571379
|
2.108
|
|
CD28
|
CD28 molecule
|